Reshaping data with tidyr
Last updated on 2022-09-07 | Edit this page
Estimated time 66 minutes
Overview
Questions
- How can I make my data into a longer format?
- How can I get my data into a wider format?
Objectives
- Use
pivot_longer()to reshape data into a longer format - Use
pivot_wider()to reshape data into a wider format
Motivation
Data come in a myriad of different shapes, and talking about data set can often become confusing as people are used to data being in different formats, and they call these formats different things. In the tidyverse, “tidy” data is a very opinionated term so that we can all talk about data with more common ground.
The goal of the tidyr package is to help you create tidy data.
Tidy data is data where:
- Every column is variable.
- Every row is an observation.
- Every cell is a single value.
Tidy data describes a standard way of storing data that is used wherever possible throughout the tidyverse. If you ensure that your data is tidy, you’ll spend less time fighting with the tools and more time working on your analysis. Learn more about tidy data in vignette("tidy-data").
Tall/long vs. wide data
Tall (or long) data are considered “tidy”, in that they adhere to the three tidy-data principles
Wide data are not necessarily “messy”, but have a shape less ideal for easy handling in the tidyverse
Example in longitudinal data design:
- wide data: each participant has a single row of data, with all longitudinal observations in separate columns
- tall data: a participant has as many rows as longitudinal time points, with measures in separate columns
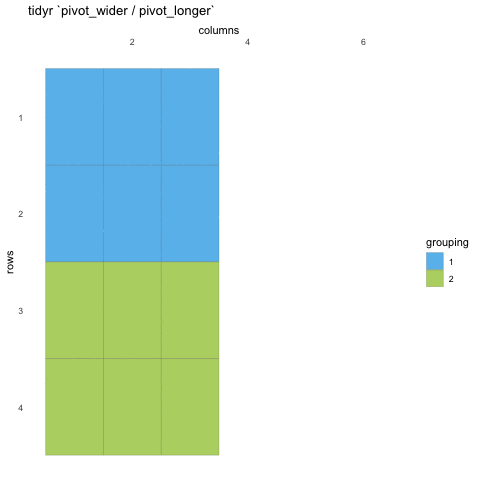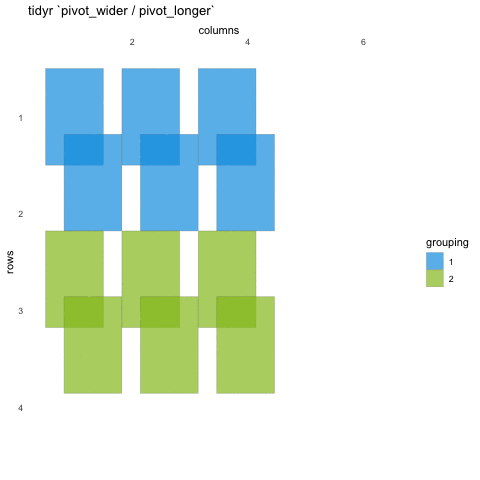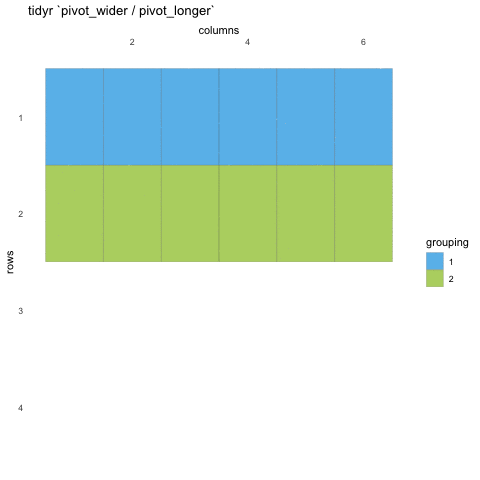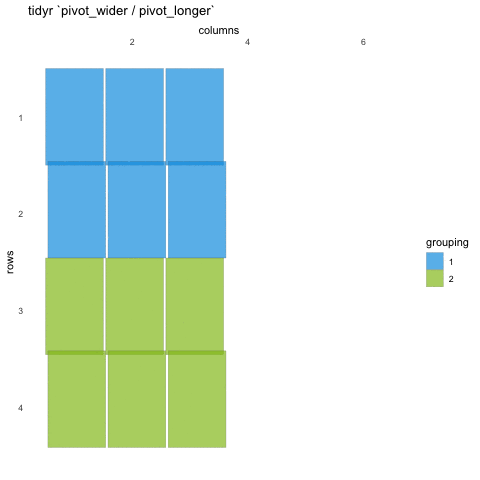
Creating longer data
Let us first talk about creating longer data. In most cases, you will encounter data that is in wide format, this is what is often taught in many disciplines and also necessary to run certain analyses in statistical programs like SPSS. In R, and specifically the tidyverse, working on long data has clear advantages, which we wil be exploring here while we also do the transformations.
As before, we need to start off by making sure we have the tidyverse package loaded, and the penguins dataset ready at hand.
In tidyverse, there is a single function to create longer data sets, called pivot_longer. Those of you who might have some prior experience with tidyverse, or you might encounter it when googling for help, might have seen the gather function. This is an older function of similar capabilities which we will not cover here, as the pivot_longer function supersedes it.
R
penguins |>
pivot_longer(contains("_"))
OUTPUT
# A tibble: 1,376 × 6
species island sex year name value
<fct> <fct> <fct> <int> <chr> <dbl>
1 Adelie Torgersen male 2007 bill_length_mm 39.1
2 Adelie Torgersen male 2007 bill_depth_mm 18.7
3 Adelie Torgersen male 2007 flipper_length_mm 181
4 Adelie Torgersen male 2007 body_mass_g 3750
5 Adelie Torgersen female 2007 bill_length_mm 39.5
6 Adelie Torgersen female 2007 bill_depth_mm 17.4
7 Adelie Torgersen female 2007 flipper_length_mm 186
8 Adelie Torgersen female 2007 body_mass_g 3800
9 Adelie Torgersen female 2007 bill_length_mm 40.3
10 Adelie Torgersen female 2007 bill_depth_mm 18
# … with 1,366 more rows
# ℹ Use `print(n = ...)` to see more rowspivot_longer takes tidy-select column arguments, so it is easy to grab all the columns you are after. Here, we are pivoting longer all columns that contain an underscore. And what happens? We now have less columns, but also two new columns we did not have before! In the name column, all our previous columns names are, one after the other. And in the value column, all the cell values for the observations! So before, the data was wider, in that each of the columns with _ had their own column, while now, they are all collected into two columns instead of 4.
Why would we want to do that? Well, perhaps we want to plot all the variables in a single ggplot call? Now that the measurement types are collected in these two ways, we can facet over the name column to create a sub-plot per measurement type!
R
penguins |>
pivot_longer(contains("_")) |>
ggplot(aes(y = value,
x = species,
fill = species)) +
geom_boxplot() +
facet_wrap(~name, scales = "free_y")
WARNING
Warning: Removed 8 rows containing non-finite values (stat_boxplot).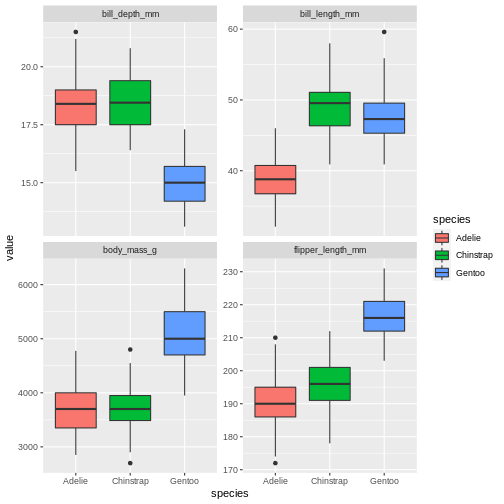
That’s pretty neat. By pivoting the data into this longer shape we are able to create sub-plots for all measurements easily with the same ggplot call and have them consistent, and nicely aligned. This longer format is also great for summaries, which we will be covering tomorrow.
R
penguins |>
pivot_longer(ends_with("mm"))
OUTPUT
# A tibble: 1,032 × 7
species island body_mass_g sex year name value
<fct> <fct> <int> <fct> <int> <chr> <dbl>
1 Adelie Torgersen 3750 male 2007 bill_length_mm 39.1
2 Adelie Torgersen 3750 male 2007 bill_depth_mm 18.7
3 Adelie Torgersen 3750 male 2007 flipper_length_mm 181
4 Adelie Torgersen 3800 female 2007 bill_length_mm 39.5
5 Adelie Torgersen 3800 female 2007 bill_depth_mm 17.4
6 Adelie Torgersen 3800 female 2007 flipper_length_mm 186
7 Adelie Torgersen 3250 female 2007 bill_length_mm 40.3
8 Adelie Torgersen 3250 female 2007 bill_depth_mm 18
9 Adelie Torgersen 3250 female 2007 flipper_length_mm 195
10 Adelie Torgersen NA <NA> 2007 bill_length_mm NA
# … with 1,022 more rows
# ℹ Use `print(n = ...)` to see more rowsR
penguins |>
pivot_longer(starts_with("bill"))
OUTPUT
# A tibble: 688 × 8
species island flipper_length_mm body_mass_g sex year name value
<fct> <fct> <int> <int> <fct> <int> <chr> <dbl>
1 Adelie Torgersen 181 3750 male 2007 bill_leng… 39.1
2 Adelie Torgersen 181 3750 male 2007 bill_dept… 18.7
3 Adelie Torgersen 186 3800 female 2007 bill_leng… 39.5
4 Adelie Torgersen 186 3800 female 2007 bill_dept… 17.4
5 Adelie Torgersen 195 3250 female 2007 bill_leng… 40.3
6 Adelie Torgersen 195 3250 female 2007 bill_dept… 18
7 Adelie Torgersen NA NA <NA> 2007 bill_leng… NA
8 Adelie Torgersen NA NA <NA> 2007 bill_dept… NA
9 Adelie Torgersen 193 3450 female 2007 bill_leng… 36.7
10 Adelie Torgersen 193 3450 female 2007 bill_dept… 19.3
# … with 678 more rows
# ℹ Use `print(n = ...)` to see more rowsR
penguins |>
pivot_longer(where(is.numeric))
OUTPUT
# A tibble: 1,720 × 5
species island sex name value
<fct> <fct> <fct> <chr> <dbl>
1 Adelie Torgersen male bill_length_mm 39.1
2 Adelie Torgersen male bill_depth_mm 18.7
3 Adelie Torgersen male flipper_length_mm 181
4 Adelie Torgersen male body_mass_g 3750
5 Adelie Torgersen male year 2007
6 Adelie Torgersen female bill_length_mm 39.5
7 Adelie Torgersen female bill_depth_mm 17.4
8 Adelie Torgersen female flipper_length_mm 186
9 Adelie Torgersen female body_mass_g 3800
10 Adelie Torgersen female year 2007
# … with 1,710 more rows
# ℹ Use `print(n = ...)` to see more rowsAltering names during pivots
While often you can get away with leaving the default naming of the two columns as is, especially if you are just doing something quick like making a plot, most times you will likely want to control the names of your two new columns.
R
penguins |>
pivot_longer(contains("_"),
names_to = "columns",
values_to = "content")
OUTPUT
# A tibble: 1,376 × 6
species island sex year columns content
<fct> <fct> <fct> <int> <chr> <dbl>
1 Adelie Torgersen male 2007 bill_length_mm 39.1
2 Adelie Torgersen male 2007 bill_depth_mm 18.7
3 Adelie Torgersen male 2007 flipper_length_mm 181
4 Adelie Torgersen male 2007 body_mass_g 3750
5 Adelie Torgersen female 2007 bill_length_mm 39.5
6 Adelie Torgersen female 2007 bill_depth_mm 17.4
7 Adelie Torgersen female 2007 flipper_length_mm 186
8 Adelie Torgersen female 2007 body_mass_g 3800
9 Adelie Torgersen female 2007 bill_length_mm 40.3
10 Adelie Torgersen female 2007 bill_depth_mm 18
# … with 1,366 more rows
# ℹ Use `print(n = ...)` to see more rowsHere, we change the “names” to “columns” and “values” to “content”. The pivot defaults are usually quite sensible, making it clear what is the column names and what are the cell values. But English might not be your working language or you might find something more obvious for your self.
But we have even more power in the renaming of columns. Pivots actually have quite a lot of options, making it possible for us to create outputs looking just like we want. Notice how the names of the columns we pivoted follow a specific structure. First is the name of the body part, then the type of measurement, then the unit of the measurement. This clear logic we can use to our advantage.
R
penguins |>
pivot_longer(contains("_"),
names_to = c("part", "measure" , "unit"),
names_sep = "_")
OUTPUT
# A tibble: 1,376 × 8
species island sex year part measure unit value
<fct> <fct> <fct> <int> <chr> <chr> <chr> <dbl>
1 Adelie Torgersen male 2007 bill length mm 39.1
2 Adelie Torgersen male 2007 bill depth mm 18.7
3 Adelie Torgersen male 2007 flipper length mm 181
4 Adelie Torgersen male 2007 body mass g 3750
5 Adelie Torgersen female 2007 bill length mm 39.5
6 Adelie Torgersen female 2007 bill depth mm 17.4
7 Adelie Torgersen female 2007 flipper length mm 186
8 Adelie Torgersen female 2007 body mass g 3800
9 Adelie Torgersen female 2007 bill length mm 40.3
10 Adelie Torgersen female 2007 bill depth mm 18
# … with 1,366 more rows
# ℹ Use `print(n = ...)` to see more rowsnow, the pivot gave us 4 columns in stead of two! We told pivot that the column name could be split into the columns “part”, “measure” and “unit”, and that these were separated by underscore. Again we see how great consistent and logical naming of columns can be such a great help when working with data!
R
penguins |>
pivot_longer(starts_with("bill"),
names_to = c("part", "measure" , "unit"),
names_sep = "_")
OUTPUT
# A tibble: 688 × 10
species island flipper_len…¹ body_…² sex year part measure unit value
<fct> <fct> <int> <int> <fct> <int> <chr> <chr> <chr> <dbl>
1 Adelie Torgersen 181 3750 male 2007 bill length mm 39.1
2 Adelie Torgersen 181 3750 male 2007 bill depth mm 18.7
3 Adelie Torgersen 186 3800 fema… 2007 bill length mm 39.5
4 Adelie Torgersen 186 3800 fema… 2007 bill depth mm 17.4
5 Adelie Torgersen 195 3250 fema… 2007 bill length mm 40.3
6 Adelie Torgersen 195 3250 fema… 2007 bill depth mm 18
7 Adelie Torgersen NA NA <NA> 2007 bill length mm NA
8 Adelie Torgersen NA NA <NA> 2007 bill depth mm NA
9 Adelie Torgersen 193 3450 fema… 2007 bill length mm 36.7
10 Adelie Torgersen 193 3450 fema… 2007 bill depth mm 19.3
# … with 678 more rows, and abbreviated variable names ¹flipper_length_mm,
# ²body_mass_g
# ℹ Use `print(n = ...)` to see more rowsR
penguins |>
pivot_longer(starts_with("bill"),
names_prefix = "bill_")
OUTPUT
# A tibble: 688 × 8
species island flipper_length_mm body_mass_g sex year name value
<fct> <fct> <int> <int> <fct> <int> <chr> <dbl>
1 Adelie Torgersen 181 3750 male 2007 length_mm 39.1
2 Adelie Torgersen 181 3750 male 2007 depth_mm 18.7
3 Adelie Torgersen 186 3800 female 2007 length_mm 39.5
4 Adelie Torgersen 186 3800 female 2007 depth_mm 17.4
5 Adelie Torgersen 195 3250 female 2007 length_mm 40.3
6 Adelie Torgersen 195 3250 female 2007 depth_mm 18
7 Adelie Torgersen NA NA <NA> 2007 length_mm NA
8 Adelie Torgersen NA NA <NA> 2007 depth_mm NA
9 Adelie Torgersen 193 3450 female 2007 length_mm 36.7
10 Adelie Torgersen 193 3450 female 2007 depth_mm 19.3
# … with 678 more rows
# ℹ Use `print(n = ...)` to see more rowsR
penguins |>
pivot_longer(starts_with("bill"),
names_prefix = "bill_",
names_to = c("bill_measure" , "unit"),
names_sep = "_")
OUTPUT
# A tibble: 688 × 9
species island flipper_length_mm body_ma…¹ sex year bill_…² unit value
<fct> <fct> <int> <int> <fct> <int> <chr> <chr> <dbl>
1 Adelie Torgersen 181 3750 male 2007 length mm 39.1
2 Adelie Torgersen 181 3750 male 2007 depth mm 18.7
3 Adelie Torgersen 186 3800 fema… 2007 length mm 39.5
4 Adelie Torgersen 186 3800 fema… 2007 depth mm 17.4
5 Adelie Torgersen 195 3250 fema… 2007 length mm 40.3
6 Adelie Torgersen 195 3250 fema… 2007 depth mm 18
7 Adelie Torgersen NA NA <NA> 2007 length mm NA
8 Adelie Torgersen NA NA <NA> 2007 depth mm NA
9 Adelie Torgersen 193 3450 fema… 2007 length mm 36.7
10 Adelie Torgersen 193 3450 fema… 2007 depth mm 19.3
# … with 678 more rows, and abbreviated variable names ¹body_mass_g,
# ²bill_measure
# ℹ Use `print(n = ...)` to see more rowsCleaning up values during pivots.
When pivoting, it is common that quite some NA values appear in the values column. We can remove these immediately by making the argument values_drop_na be TRUE
R
penguins |>
pivot_longer(starts_with("bill"),
values_drop_na = TRUE)
OUTPUT
# A tibble: 684 × 8
species island flipper_length_mm body_mass_g sex year name value
<fct> <fct> <int> <int> <fct> <int> <chr> <dbl>
1 Adelie Torgersen 181 3750 male 2007 bill_leng… 39.1
2 Adelie Torgersen 181 3750 male 2007 bill_dept… 18.7
3 Adelie Torgersen 186 3800 female 2007 bill_leng… 39.5
4 Adelie Torgersen 186 3800 female 2007 bill_dept… 17.4
5 Adelie Torgersen 195 3250 female 2007 bill_leng… 40.3
6 Adelie Torgersen 195 3250 female 2007 bill_dept… 18
7 Adelie Torgersen 193 3450 female 2007 bill_leng… 36.7
8 Adelie Torgersen 193 3450 female 2007 bill_dept… 19.3
9 Adelie Torgersen 190 3650 male 2007 bill_leng… 39.3
10 Adelie Torgersen 190 3650 male 2007 bill_dept… 20.6
# … with 674 more rows
# ℹ Use `print(n = ...)` to see more rowsThis extra argument will ensure that all NA values in the value column are removed. This is some times convenient as we might move on to analyses etc of the data, which often are made more complicated (or impossible) when there is missing data.
We should put everything together and create a new object that is our long formatted penguin data set.
R
penguins_long <- penguins |>
pivot_longer(contains("_"),
names_to = c("part", "measure" , "unit"),
names_sep = "_",
values_drop_na = TRUE)
penguins_long
OUTPUT
# A tibble: 1,368 × 8
species island sex year part measure unit value
<fct> <fct> <fct> <int> <chr> <chr> <chr> <dbl>
1 Adelie Torgersen male 2007 bill length mm 39.1
2 Adelie Torgersen male 2007 bill depth mm 18.7
3 Adelie Torgersen male 2007 flipper length mm 181
4 Adelie Torgersen male 2007 body mass g 3750
5 Adelie Torgersen female 2007 bill length mm 39.5
6 Adelie Torgersen female 2007 bill depth mm 17.4
7 Adelie Torgersen female 2007 flipper length mm 186
8 Adelie Torgersen female 2007 body mass g 3800
9 Adelie Torgersen female 2007 bill length mm 40.3
10 Adelie Torgersen female 2007 bill depth mm 18
# … with 1,358 more rows
# ℹ Use `print(n = ...)` to see more rowsPivoting data wider
While long data formats are ideal when you are working in the tidyverse, you might encounter packages or pipelines in R that require wide-format data. Knowing how to transform a long data set into wide is just as important a knowing how to go from wide to long. You will also experience that this skill can be convenient when creating data summaries tomorrow.
Before we start using the penguins_longer dataset we made, let us make another simpler longer data set, for the first look a the pivor wider function.
R
penguins_long_simple <- penguins |>
pivot_longer(contains("_"))
penguins_long_simple
OUTPUT
# A tibble: 1,376 × 6
species island sex year name value
<fct> <fct> <fct> <int> <chr> <dbl>
1 Adelie Torgersen male 2007 bill_length_mm 39.1
2 Adelie Torgersen male 2007 bill_depth_mm 18.7
3 Adelie Torgersen male 2007 flipper_length_mm 181
4 Adelie Torgersen male 2007 body_mass_g 3750
5 Adelie Torgersen female 2007 bill_length_mm 39.5
6 Adelie Torgersen female 2007 bill_depth_mm 17.4
7 Adelie Torgersen female 2007 flipper_length_mm 186
8 Adelie Torgersen female 2007 body_mass_g 3800
9 Adelie Torgersen female 2007 bill_length_mm 40.3
10 Adelie Torgersen female 2007 bill_depth_mm 18
# … with 1,366 more rows
# ℹ Use `print(n = ...)` to see more rowspenguins_long_simple now contains the lover penguins dataset, with column names in the “name” column, and values in the “value” column.
If we want to make this wider again we can try the following:
R
penguins_long_simple |>
pivot_wider(names_from = name,
values_from = value)
WARNING
Warning: Values from `value` are not uniquely identified; output will contain list-cols.
* Use `values_fn = list` to suppress this warning.
* Use `values_fn = {summary_fun}` to summarise duplicates.
* Use the following dplyr code to identify duplicates.
{data} %>%
dplyr::group_by(species, island, sex, year, name) %>%
dplyr::summarise(n = dplyr::n(), .groups = "drop") %>%
dplyr::filter(n > 1L)OUTPUT
# A tibble: 35 × 8
species island sex year bill_length_mm bill_depth_mm flipper…¹ body_…²
<fct> <fct> <fct> <int> <list> <list> <list> <list>
1 Adelie Torgersen male 2007 <dbl [7]> <dbl [7]> <dbl [7]> <dbl>
2 Adelie Torgersen female 2007 <dbl [8]> <dbl [8]> <dbl [8]> <dbl>
3 Adelie Torgersen <NA> 2007 <dbl [5]> <dbl [5]> <dbl [5]> <dbl>
4 Adelie Biscoe female 2007 <dbl [5]> <dbl [5]> <dbl [5]> <dbl>
5 Adelie Biscoe male 2007 <dbl [5]> <dbl [5]> <dbl [5]> <dbl>
6 Adelie Dream female 2007 <dbl [9]> <dbl [9]> <dbl [9]> <dbl>
7 Adelie Dream male 2007 <dbl [10]> <dbl [10]> <dbl> <dbl>
8 Adelie Dream <NA> 2007 <dbl [1]> <dbl [1]> <dbl [1]> <dbl>
9 Adelie Biscoe female 2008 <dbl [9]> <dbl [9]> <dbl [9]> <dbl>
10 Adelie Biscoe male 2008 <dbl [9]> <dbl [9]> <dbl [9]> <dbl>
# … with 25 more rows, and abbreviated variable names ¹flipper_length_mm,
# ²body_mass_g
# ℹ Use `print(n = ...)` to see more rowsok what is happening here? It does not at all look as we expected! Our columns have something very weird in them, with this strange <dbl [7]> thing, what does that mean? Lets look at the warning message our code gave us and see if we can figure it out. Values are not uniquely identified; output will contain list-cols. We are being told the pivot wider cannot uniquely identify the observations, and so cannot place a single value into the columns. Is returning lists of values.
yikes! That’s super annoying. Let’s go back to our penguins data set and see if we can do something to help.
R
penguins
OUTPUT
# A tibble: 344 × 8
species island bill_length_mm bill_depth_mm flipper_…¹ body_…² sex year
<fct> <fct> <dbl> <dbl> <int> <int> <fct> <int>
1 Adelie Torgersen 39.1 18.7 181 3750 male 2007
2 Adelie Torgersen 39.5 17.4 186 3800 fema… 2007
3 Adelie Torgersen 40.3 18 195 3250 fema… 2007
4 Adelie Torgersen NA NA NA NA <NA> 2007
5 Adelie Torgersen 36.7 19.3 193 3450 fema… 2007
6 Adelie Torgersen 39.3 20.6 190 3650 male 2007
7 Adelie Torgersen 38.9 17.8 181 3625 fema… 2007
8 Adelie Torgersen 39.2 19.6 195 4675 male 2007
9 Adelie Torgersen 34.1 18.1 193 3475 <NA> 2007
10 Adelie Torgersen 42 20.2 190 4250 <NA> 2007
# … with 334 more rows, and abbreviated variable names ¹flipper_length_mm,
# ²body_mass_g
# ℹ Use `print(n = ...)` to see more rowsHave you noticed that there is no column that uniquely identifies an observation? Other than each observation being on its own row, we have nothing to make sure that we can identify which observations belong together once we make the data long. As long as they are in the original format, this is ok, but once we pivoted the data longer, we lost the ability to identify which rows of observations belong together.
We can remedy that by adding row numbers to the original data before we pivot. The row_number() function is great for this. By doing a mutate adding the row number to the data set, we should then have a clear variable identifying each observation.
R
penguins_long_simple <- penguins |>
mutate(sample = row_number()) |>
pivot_longer(contains("_"))
penguins_long_simple
OUTPUT
# A tibble: 1,376 × 7
species island sex year sample name value
<fct> <fct> <fct> <int> <int> <chr> <dbl>
1 Adelie Torgersen male 2007 1 bill_length_mm 39.1
2 Adelie Torgersen male 2007 1 bill_depth_mm 18.7
3 Adelie Torgersen male 2007 1 flipper_length_mm 181
4 Adelie Torgersen male 2007 1 body_mass_g 3750
5 Adelie Torgersen female 2007 2 bill_length_mm 39.5
6 Adelie Torgersen female 2007 2 bill_depth_mm 17.4
7 Adelie Torgersen female 2007 2 flipper_length_mm 186
8 Adelie Torgersen female 2007 2 body_mass_g 3800
9 Adelie Torgersen female 2007 3 bill_length_mm 40.3
10 Adelie Torgersen female 2007 3 bill_depth_mm 18
# … with 1,366 more rows
# ℹ Use `print(n = ...)` to see more rowsNotice now that in the sample column, the numbers repeat several rows. Where sample equals 1, all those are observations from the first row of data in the original penguins data set! Let us try to pivot that wider again.
R
penguins_long_simple |>
pivot_wider(names_from = name,
values_from = value)
OUTPUT
# A tibble: 344 × 9
species island sex year sample bill_length_mm bill_d…¹ flipp…² body_…³
<fct> <fct> <fct> <int> <int> <dbl> <dbl> <dbl> <dbl>
1 Adelie Torgersen male 2007 1 39.1 18.7 181 3750
2 Adelie Torgersen female 2007 2 39.5 17.4 186 3800
3 Adelie Torgersen female 2007 3 40.3 18 195 3250
4 Adelie Torgersen <NA> 2007 4 NA NA NA NA
5 Adelie Torgersen female 2007 5 36.7 19.3 193 3450
6 Adelie Torgersen male 2007 6 39.3 20.6 190 3650
7 Adelie Torgersen female 2007 7 38.9 17.8 181 3625
8 Adelie Torgersen male 2007 8 39.2 19.6 195 4675
9 Adelie Torgersen <NA> 2007 9 34.1 18.1 193 3475
10 Adelie Torgersen <NA> 2007 10 42 20.2 190 4250
# … with 334 more rows, and abbreviated variable names ¹bill_depth_mm,
# ²flipper_length_mm, ³body_mass_g
# ℹ Use `print(n = ...)` to see more rowsAnd now it worked! Now, the remaining columns were able to uniquely identify which observations belonged together. And the data looks just like the original penguins data set now, with the addition of the sample column, and the columns being slightly rearranged.
Pivoting wider with more arguments
We should re-create our penguins long data set, to make sure we don’t have this problem again.
R
penguins_long <- penguins |>
mutate(sample = row_number()) |>
pivot_longer(contains("_"),
names_to = c("part", "measure" , "unit"),
names_sep = "_",
values_drop_na = TRUE)
penguins_long
OUTPUT
# A tibble: 1,368 × 9
species island sex year sample part measure unit value
<fct> <fct> <fct> <int> <int> <chr> <chr> <chr> <dbl>
1 Adelie Torgersen male 2007 1 bill length mm 39.1
2 Adelie Torgersen male 2007 1 bill depth mm 18.7
3 Adelie Torgersen male 2007 1 flipper length mm 181
4 Adelie Torgersen male 2007 1 body mass g 3750
5 Adelie Torgersen female 2007 2 bill length mm 39.5
6 Adelie Torgersen female 2007 2 bill depth mm 17.4
7 Adelie Torgersen female 2007 2 flipper length mm 186
8 Adelie Torgersen female 2007 2 body mass g 3800
9 Adelie Torgersen female 2007 3 bill length mm 40.3
10 Adelie Torgersen female 2007 3 bill depth mm 18
# … with 1,358 more rows
# ℹ Use `print(n = ...)` to see more rowsMuch as the first example of pivot_longer, pivot_wider in its simplest form is relatively straight forward. But your penguins long data set is much more complex. The column names are split into several columns, how do we fix that? Like pivot_longer, pivot_wider has arguments that will let us get back to the original state, with much of the same syntax as with pivot_longer!
R
penguins_long |>
pivot_wider(names_from = c("part", "measure", "unit"),
names_sep = "_",
values_from = value)
OUTPUT
# A tibble: 342 × 9
species island sex year sample bill_length_mm bill_d…¹ flipp…² body_…³
<fct> <fct> <fct> <int> <int> <dbl> <dbl> <dbl> <dbl>
1 Adelie Torgersen male 2007 1 39.1 18.7 181 3750
2 Adelie Torgersen female 2007 2 39.5 17.4 186 3800
3 Adelie Torgersen female 2007 3 40.3 18 195 3250
4 Adelie Torgersen female 2007 5 36.7 19.3 193 3450
5 Adelie Torgersen male 2007 6 39.3 20.6 190 3650
6 Adelie Torgersen female 2007 7 38.9 17.8 181 3625
7 Adelie Torgersen male 2007 8 39.2 19.6 195 4675
8 Adelie Torgersen <NA> 2007 9 34.1 18.1 193 3475
9 Adelie Torgersen <NA> 2007 10 42 20.2 190 4250
10 Adelie Torgersen <NA> 2007 11 37.8 17.1 186 3300
# … with 332 more rows, and abbreviated variable names ¹bill_depth_mm,
# ²flipper_length_mm, ³body_mass_g
# ℹ Use `print(n = ...)` to see more rowsThose arguments and inputs should be familiar to the call from pivot_longer. So we are lucky that if you understand one of them, it is easier to understand the other.
Wrap up
We have been exploring how to pivot data into longer and wider shapes. Pivoting is a vital part of the “tidyverse”-way, and very powerful tool once you get used to it. We will see pivots in action more tomorrow as we create summaries and play around with combining all the things we have been exploring.
