Subsetting data with dplyr
Last updated on 2022-09-07 | Edit this page
Overview
Questions
- How can I subset the number of columns in my data set?
- How can I reduce the number of rows in my data set?
Objectives
- Use
select()to reduce columns - Use tidyselectors like
starts_with()withinselect()to reduce columns - Use
filter()to reduce rows - Understand common logical operations using
filter()
Motivation
In many cases, we are working with data sets that contain more data than we need, or we want to inspect certain parts of the data set before we continue. Subsetting data sets can be challenging in base R, because there is a fair bit of repetition. This can make code difficult to readn and understand.
The {dplyr} package
The {dplyr} package provides a number of very useful functions for manipulating data sets in a way that will reduce the probability of making errors, and even save you some typing time. As an added bonus, you might even find the {dplyr} grammar easier to read.
We’re going to cover 6 of the most commonly used functions as well as using pipes (|>) to combine them.
-
select()(covered in this session) -
filter()(covered in this session) -
arrange()(covered in this session) -
mutate()(covered in next session) -
group_by()(covered in Day 2 session) -
summarize()(covered in Day 2 session)
Selecting columns
Let us first talk about selecting columns. In {dplyr}, the function name for selecting columns is select()! Most {tidyverse} function names for functions are inspired by English grammar, which will help us when we are writing our code.
We first need to make sure we have the tidyverse loaded and the penguins data set at hand.
R
library(tidyverse)
penguins <- read_csv("data/penguins.csv")
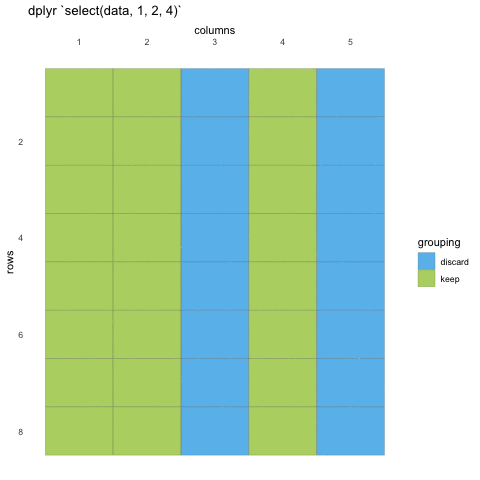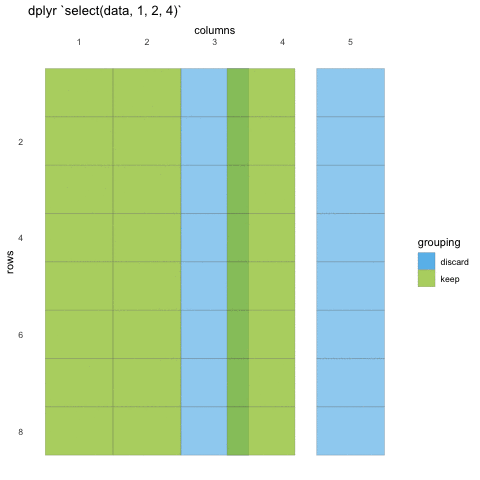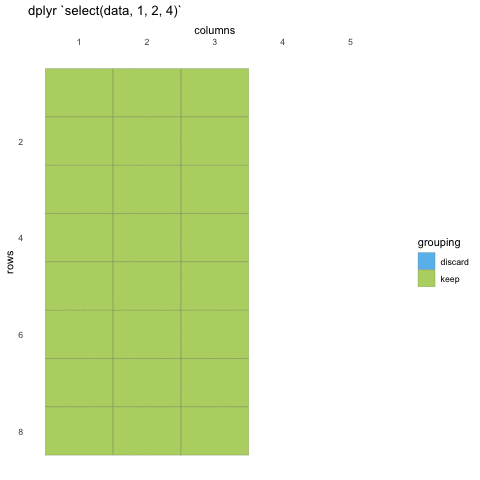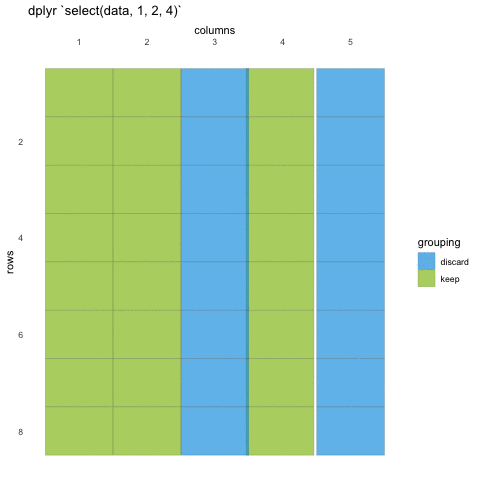
To select data, we must first tell select which data set we are selecting from, and then give it our selection. Here, we are asking R to select() from the penguins data set the island, species and sex columns
R
select(penguins, island, species, sex)
OUTPUT
# A tibble: 344 × 3
island species sex
<fct> <fct> <fct>
1 Torgersen Adelie male
2 Torgersen Adelie female
3 Torgersen Adelie female
4 Torgersen Adelie <NA>
5 Torgersen Adelie female
6 Torgersen Adelie male
7 Torgersen Adelie female
8 Torgersen Adelie male
9 Torgersen Adelie <NA>
10 Torgersen Adelie <NA>
# … with 334 more rows
# ℹ Use `print(n = ...)` to see more rowsWhen we use select() we don’t need to use quotations, we write in the names directly. We can also use the numeric indexes for the column, if we are 100% certain of the order of the columns:
R
select(penguins, 1:3, 6)
OUTPUT
# A tibble: 344 × 4
species island bill_length_mm body_mass_g
<fct> <fct> <dbl> <int>
1 Adelie Torgersen 39.1 3750
2 Adelie Torgersen 39.5 3800
3 Adelie Torgersen 40.3 3250
4 Adelie Torgersen NA NA
5 Adelie Torgersen 36.7 3450
6 Adelie Torgersen 39.3 3650
7 Adelie Torgersen 38.9 3625
8 Adelie Torgersen 39.2 4675
9 Adelie Torgersen 34.1 3475
10 Adelie Torgersen 42 4250
# … with 334 more rows
# ℹ Use `print(n = ...)` to see more rowsIn some cases, we want to remove columns, and not necessarily state all columns we want to keep. Select also allows for this by adding a minus (-) sign in front of the column name you don’t want.
R
select(penguins, -bill_length_mm, -bill_depth_mm)
OUTPUT
# A tibble: 344 × 6
species island flipper_length_mm body_mass_g sex year
<fct> <fct> <int> <int> <fct> <int>
1 Adelie Torgersen 181 3750 male 2007
2 Adelie Torgersen 186 3800 female 2007
3 Adelie Torgersen 195 3250 female 2007
4 Adelie Torgersen NA NA <NA> 2007
5 Adelie Torgersen 193 3450 female 2007
6 Adelie Torgersen 190 3650 male 2007
7 Adelie Torgersen 181 3625 female 2007
8 Adelie Torgersen 195 4675 male 2007
9 Adelie Torgersen 193 3475 <NA> 2007
10 Adelie Torgersen 190 4250 <NA> 2007
# … with 334 more rows
# ℹ Use `print(n = ...)` to see more rowsR
select(penguins, sex, year, species)
OUTPUT
# A tibble: 344 × 3
sex year species
<fct> <int> <fct>
1 male 2007 Adelie
2 female 2007 Adelie
3 female 2007 Adelie
4 <NA> 2007 Adelie
5 female 2007 Adelie
6 male 2007 Adelie
7 female 2007 Adelie
8 male 2007 Adelie
9 <NA> 2007 Adelie
10 <NA> 2007 Adelie
# … with 334 more rows
# ℹ Use `print(n = ...)` to see more rowsR
select(penguins, species, sex, year)
OUTPUT
# A tibble: 344 × 3
species sex year
<fct> <fct> <int>
1 Adelie male 2007
2 Adelie female 2007
3 Adelie female 2007
4 Adelie <NA> 2007
5 Adelie female 2007
6 Adelie male 2007
7 Adelie female 2007
8 Adelie male 2007
9 Adelie <NA> 2007
10 Adelie <NA> 2007
# … with 334 more rows
# ℹ Use `print(n = ...)` to see more rowsselect does not only subset columns, but it can also re-arrange them. The columns appear in the order your selection is specified.
Tidy selections
These selections are quite convenient and fast! But they can be even better.
For instance, what if we want to choose all the columns with millimeter measurements? That could be quite convenient, making sure the variables we are working with have the same measurement scale.
We could of course type them all out, but the penguins data set has names that make it even easier for us, using something called tidy-selectors.
Here, we use a tidy-selector ends_with(), can you guess what it does? yes, it looks for columns that end with the string you provide it, here "mm".
R
select(penguins, ends_with("mm"))
OUTPUT
# A tibble: 344 × 3
bill_length_mm bill_depth_mm flipper_length_mm
<dbl> <dbl> <int>
1 39.1 18.7 181
2 39.5 17.4 186
3 40.3 18 195
4 NA NA NA
5 36.7 19.3 193
6 39.3 20.6 190
7 38.9 17.8 181
8 39.2 19.6 195
9 34.1 18.1 193
10 42 20.2 190
# … with 334 more rows
# ℹ Use `print(n = ...)` to see more rowsSo convenient! There are several other tidy-selectors you can choose, which you can find here, but often people resort to three specific ones:
-
ends_with()- column names ending with a character string -
starts_with()- column names starting with a character string -
contains()- column names containing a character string
If you are working with a well named data set, these functions should make your data selecting much simpler. And if you are making your own data, you can think of such convenient naming for your data, so your work can be easier for you and others.
Lets only pick the measurements of the bill, we are not so interested in the flipper. Then we might want to change to starts_with() in stead.
R
select(penguins, starts_with("bill"))
OUTPUT
# A tibble: 344 × 2
bill_length_mm bill_depth_mm
<dbl> <dbl>
1 39.1 18.7
2 39.5 17.4
3 40.3 18
4 NA NA
5 36.7 19.3
6 39.3 20.6
7 38.9 17.8
8 39.2 19.6
9 34.1 18.1
10 42 20.2
# … with 334 more rows
# ℹ Use `print(n = ...)` to see more rowsThe tidy selector can be combined with each other and other selectors. So you can build exactly the data you want!
R
select(penguins, island, species, year, starts_with("bill"))
OUTPUT
# A tibble: 344 × 5
island species year bill_length_mm bill_depth_mm
<fct> <fct> <int> <dbl> <dbl>
1 Torgersen Adelie 2007 39.1 18.7
2 Torgersen Adelie 2007 39.5 17.4
3 Torgersen Adelie 2007 40.3 18
4 Torgersen Adelie 2007 NA NA
5 Torgersen Adelie 2007 36.7 19.3
6 Torgersen Adelie 2007 39.3 20.6
7 Torgersen Adelie 2007 38.9 17.8
8 Torgersen Adelie 2007 39.2 19.6
9 Torgersen Adelie 2007 34.1 18.1
10 Torgersen Adelie 2007 42 20.2
# … with 334 more rows
# ℹ Use `print(n = ...)` to see more rowsR
select(penguins, contains("_"))
OUTPUT
# A tibble: 344 × 4
bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<dbl> <dbl> <int> <int>
1 39.1 18.7 181 3750
2 39.5 17.4 186 3800
3 40.3 18 195 3250
4 NA NA NA NA
5 36.7 19.3 193 3450
6 39.3 20.6 190 3650
7 38.9 17.8 181 3625
8 39.2 19.6 195 4675
9 34.1 18.1 193 3475
10 42 20.2 190 4250
# … with 334 more rows
# ℹ Use `print(n = ...)` to see more rowsR
select(penguins, species, sex, ends_with("mm"))
OUTPUT
# A tibble: 344 × 5
species sex bill_length_mm bill_depth_mm flipper_length_mm
<fct> <fct> <dbl> <dbl> <int>
1 Adelie male 39.1 18.7 181
2 Adelie female 39.5 17.4 186
3 Adelie female 40.3 18 195
4 Adelie <NA> NA NA NA
5 Adelie female 36.7 19.3 193
6 Adelie male 39.3 20.6 190
7 Adelie female 38.9 17.8 181
8 Adelie male 39.2 19.6 195
9 Adelie <NA> 34.1 18.1 193
10 Adelie <NA> 42 20.2 190
# … with 334 more rows
# ℹ Use `print(n = ...)` to see more rowsR
select(penguins, -starts_with("bill"))
OUTPUT
# A tibble: 344 × 6
species island flipper_length_mm body_mass_g sex year
<fct> <fct> <int> <int> <fct> <int>
1 Adelie Torgersen 181 3750 male 2007
2 Adelie Torgersen 186 3800 female 2007
3 Adelie Torgersen 195 3250 female 2007
4 Adelie Torgersen NA NA <NA> 2007
5 Adelie Torgersen 193 3450 female 2007
6 Adelie Torgersen 190 3650 male 2007
7 Adelie Torgersen 181 3625 female 2007
8 Adelie Torgersen 195 4675 male 2007
9 Adelie Torgersen 193 3475 <NA> 2007
10 Adelie Torgersen 190 4250 <NA> 2007
# … with 334 more rows
# ℹ Use `print(n = ...)` to see more rowsTidy selections with where
The last tidy-selector we’ll mention is where(). where() is a very special tidy selector, that uses logical evaluations to select the data. Let’s have a look at it in action, and see if we can explain it better that way.
Say you are running a correlation analysis. For correlations, you need all the columns in your data to be numeric, as you cannot correlate strings or categories. Going through each individual column and seeing if it is numeric is a bit of a chore. That is where where() comes in!
R
select(penguins, where(is.numeric))
OUTPUT
# A tibble: 344 × 5
bill_length_mm bill_depth_mm flipper_length_mm body_mass_g year
<dbl> <dbl> <int> <int> <int>
1 39.1 18.7 181 3750 2007
2 39.5 17.4 186 3800 2007
3 40.3 18 195 3250 2007
4 NA NA NA NA 2007
5 36.7 19.3 193 3450 2007
6 39.3 20.6 190 3650 2007
7 38.9 17.8 181 3625 2007
8 39.2 19.6 195 4675 2007
9 34.1 18.1 193 3475 2007
10 42 20.2 190 4250 2007
# … with 334 more rows
# ℹ Use `print(n = ...)` to see more rowsMagic! Let’s break that down. is.numeric() is a function in R that checks if a vector is numeric. If the vector is numeric, it returns TRUE if not it returns FALSE.
R
is.numeric(5)
OUTPUT
[1] TRUER
is.numeric("something")
OUTPUT
[1] FALSELet us look at the penguins data set again
R
penguins
OUTPUT
# A tibble: 344 × 8
species island bill_length_mm bill_depth_mm flipper_…¹ body_…² sex year
<fct> <fct> <dbl> <dbl> <int> <int> <fct> <int>
1 Adelie Torgersen 39.1 18.7 181 3750 male 2007
2 Adelie Torgersen 39.5 17.4 186 3800 fema… 2007
3 Adelie Torgersen 40.3 18 195 3250 fema… 2007
4 Adelie Torgersen NA NA NA NA <NA> 2007
5 Adelie Torgersen 36.7 19.3 193 3450 fema… 2007
6 Adelie Torgersen 39.3 20.6 190 3650 male 2007
7 Adelie Torgersen 38.9 17.8 181 3625 fema… 2007
8 Adelie Torgersen 39.2 19.6 195 4675 male 2007
9 Adelie Torgersen 34.1 18.1 193 3475 <NA> 2007
10 Adelie Torgersen 42 20.2 190 4250 <NA> 2007
# … with 334 more rows, and abbreviated variable names ¹flipper_length_mm,
# ²body_mass_g
# ℹ Use `print(n = ...)` to see more rowsThe penguins data is stored as a tibble, which is a special kind of data set in R that gives a nice print out of the data. Notice, right below the column name, there is some information in <> marks. This tells us the class of the columns. Species and island are factors, while bill columns are “double” which is a decimal numeric class.
where() goes through all the columns and checks if they are numeric, and returns the ones that are.
R
select(penguins, where(is.numeric))
OUTPUT
# A tibble: 344 × 5
bill_length_mm bill_depth_mm flipper_length_mm body_mass_g year
<dbl> <dbl> <int> <int> <int>
1 39.1 18.7 181 3750 2007
2 39.5 17.4 186 3800 2007
3 40.3 18 195 3250 2007
4 NA NA NA NA 2007
5 36.7 19.3 193 3450 2007
6 39.3 20.6 190 3650 2007
7 38.9 17.8 181 3625 2007
8 39.2 19.6 195 4675 2007
9 34.1 18.1 193 3475 2007
10 42 20.2 190 4250 2007
# … with 334 more rows
# ℹ Use `print(n = ...)` to see more rowsR
select(penguins, where(is.factor))
OUTPUT
# A tibble: 344 × 3
species island sex
<fct> <fct> <fct>
1 Adelie Torgersen male
2 Adelie Torgersen female
3 Adelie Torgersen female
4 Adelie Torgersen <NA>
5 Adelie Torgersen female
6 Adelie Torgersen male
7 Adelie Torgersen female
8 Adelie Torgersen male
9 Adelie Torgersen <NA>
10 Adelie Torgersen <NA>
# … with 334 more rows
# ℹ Use `print(n = ...)` to see more rowsR
select(penguins, island, species, where(is.numeric))
OUTPUT
# A tibble: 344 × 7
island species bill_length_mm bill_depth_mm flipper_length…¹ body_…² year
<fct> <fct> <dbl> <dbl> <int> <int> <int>
1 Torgersen Adelie 39.1 18.7 181 3750 2007
2 Torgersen Adelie 39.5 17.4 186 3800 2007
3 Torgersen Adelie 40.3 18 195 3250 2007
4 Torgersen Adelie NA NA NA NA 2007
5 Torgersen Adelie 36.7 19.3 193 3450 2007
6 Torgersen Adelie 39.3 20.6 190 3650 2007
7 Torgersen Adelie 38.9 17.8 181 3625 2007
8 Torgersen Adelie 39.2 19.6 195 4675 2007
9 Torgersen Adelie 34.1 18.1 193 3475 2007
10 Torgersen Adelie 42 20.2 190 4250 2007
# … with 334 more rows, and abbreviated variable names ¹flipper_length_mm,
# ²body_mass_g
# ℹ Use `print(n = ...)` to see more rowsFiltering rows
Now that we know how to select the columns we want, we should take a look at how we filter the rows. Row filtering is done with the function filter(), which takes statements that can be evaluated to TRUE or FALSE.
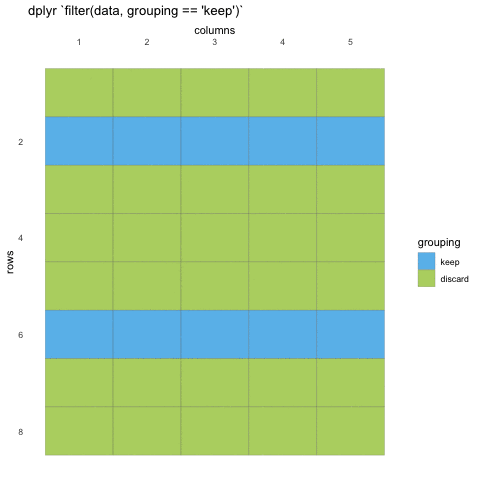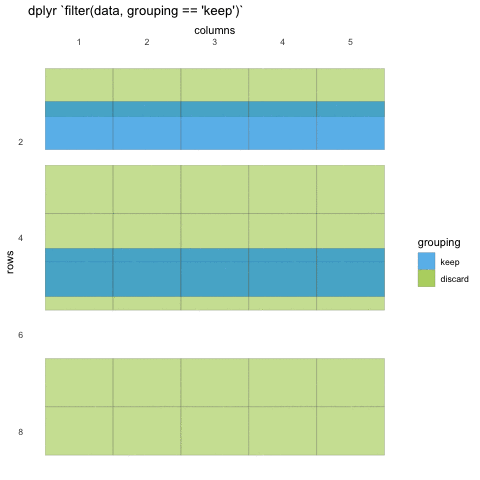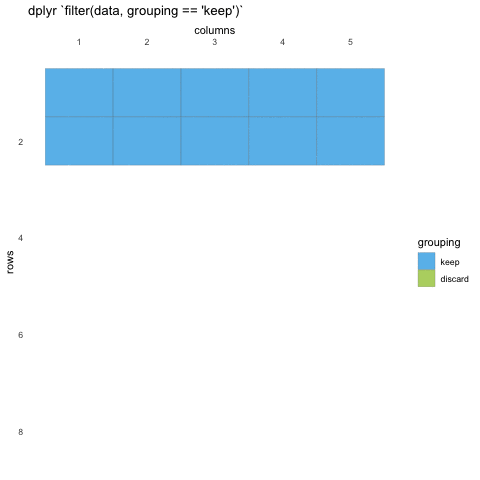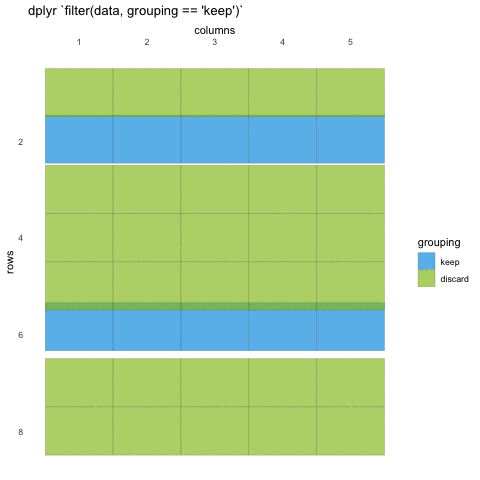
What do we mean with statements that can be evaluated to TRUE or FALSE? In the example with where() we used the is.numeric function to evaluate if the columns where numeric or not. We will be doing the same for rows!
Now, using is.numeric on a row won’t help, because every row-value in a column will be of the same type, that is how the data set works. All values in a column must be of the same type.
So what can we do? Well, we can check if the values meet certain criteria or not. Like values being above 20, or factors being a specific factor.
R
filter(penguins, body_mass_g < 3000)
OUTPUT
# A tibble: 9 × 8
species island bill_length_mm bill_depth_mm flipper…¹ body_…² sex year
<fct> <fct> <dbl> <dbl> <int> <int> <fct> <int>
1 Adelie Dream 37.5 18.9 179 2975 <NA> 2007
2 Adelie Biscoe 34.5 18.1 187 2900 fema… 2008
3 Adelie Biscoe 36.5 16.6 181 2850 fema… 2008
4 Adelie Biscoe 36.4 17.1 184 2850 fema… 2008
5 Adelie Dream 33.1 16.1 178 2900 fema… 2008
6 Adelie Biscoe 37.9 18.6 193 2925 fema… 2009
7 Adelie Torgersen 38.6 17 188 2900 fema… 2009
8 Chinstrap Dream 43.2 16.6 187 2900 fema… 2007
9 Chinstrap Dream 46.9 16.6 192 2700 fema… 2008
# … with abbreviated variable names ¹flipper_length_mm, ²body_mass_gHere, we’ve filtered so that we only have observations where the body mass was less than 3 kilos. We can also filter for specific values, but beware! you must use double equals (==) for comparisons, as single equals (=) are for argument names in functions.
R
filter(penguins, body_mass_g == 2900)
OUTPUT
# A tibble: 4 × 8
species island bill_length_mm bill_depth_mm flipper…¹ body_…² sex year
<fct> <fct> <dbl> <dbl> <int> <int> <fct> <int>
1 Adelie Biscoe 34.5 18.1 187 2900 fema… 2008
2 Adelie Dream 33.1 16.1 178 2900 fema… 2008
3 Adelie Torgersen 38.6 17 188 2900 fema… 2009
4 Chinstrap Dream 43.2 16.6 187 2900 fema… 2007
# … with abbreviated variable names ¹flipper_length_mm, ²body_mass_gWhat is happening, is that R will check if the values in body_mass_g are the same as 2900 (TRUE) or not (FALSE), and will do this for every row in the data set. Then at the end, it will discard all those that are FALSE, and keep those that are TRUE.
R
filter(penguins, island == "Dream")
OUTPUT
# A tibble: 124 × 8
species island bill_length_mm bill_depth_mm flipper_len…¹ body_…² sex year
<fct> <fct> <dbl> <dbl> <int> <int> <fct> <int>
1 Adelie Dream 39.5 16.7 178 3250 fema… 2007
2 Adelie Dream 37.2 18.1 178 3900 male 2007
3 Adelie Dream 39.5 17.8 188 3300 fema… 2007
4 Adelie Dream 40.9 18.9 184 3900 male 2007
5 Adelie Dream 36.4 17 195 3325 fema… 2007
6 Adelie Dream 39.2 21.1 196 4150 male 2007
7 Adelie Dream 38.8 20 190 3950 male 2007
8 Adelie Dream 42.2 18.5 180 3550 fema… 2007
9 Adelie Dream 37.6 19.3 181 3300 fema… 2007
10 Adelie Dream 39.8 19.1 184 4650 male 2007
# … with 114 more rows, and abbreviated variable names ¹flipper_length_mm,
# ²body_mass_g
# ℹ Use `print(n = ...)` to see more rowsR
filter(penguins, year >= 2008)
OUTPUT
# A tibble: 234 × 8
species island bill_length_mm bill_depth_mm flipper_len…¹ body_…² sex year
<fct> <fct> <dbl> <dbl> <int> <int> <fct> <int>
1 Adelie Biscoe 39.6 17.7 186 3500 fema… 2008
2 Adelie Biscoe 40.1 18.9 188 4300 male 2008
3 Adelie Biscoe 35 17.9 190 3450 fema… 2008
4 Adelie Biscoe 42 19.5 200 4050 male 2008
5 Adelie Biscoe 34.5 18.1 187 2900 fema… 2008
6 Adelie Biscoe 41.4 18.6 191 3700 male 2008
7 Adelie Biscoe 39 17.5 186 3550 fema… 2008
8 Adelie Biscoe 40.6 18.8 193 3800 male 2008
9 Adelie Biscoe 36.5 16.6 181 2850 fema… 2008
10 Adelie Biscoe 37.6 19.1 194 3750 male 2008
# … with 224 more rows, and abbreviated variable names ¹flipper_length_mm,
# ²body_mass_g
# ℹ Use `print(n = ...)` to see more rowsMultiple filters
Many times, we will want to have several filters applied at once. What if you only want Adelie penguins that are below 3 kilos? filter() can take as many statements as you want! Combine them by adding commas (,) between each statement, and that will work as ‘and’.
R
filter(penguins,
species == "Chinstrap",
body_mass_g < 3000)
OUTPUT
# A tibble: 2 × 8
species island bill_length_mm bill_depth_mm flipper_le…¹ body_…² sex year
<fct> <fct> <dbl> <dbl> <int> <int> <fct> <int>
1 Chinstrap Dream 43.2 16.6 187 2900 fema… 2007
2 Chinstrap Dream 46.9 16.6 192 2700 fema… 2008
# … with abbreviated variable names ¹flipper_length_mm, ²body_mass_gYou can also use the & sign, which in R is the comparison character for ‘and’, like == is for ‘equals’.
R
filter(penguins,
species == "Chinstrap" &
body_mass_g < 3000)
OUTPUT
# A tibble: 2 × 8
species island bill_length_mm bill_depth_mm flipper_le…¹ body_…² sex year
<fct> <fct> <dbl> <dbl> <int> <int> <fct> <int>
1 Chinstrap Dream 43.2 16.6 187 2900 fema… 2007
2 Chinstrap Dream 46.9 16.6 192 2700 fema… 2008
# … with abbreviated variable names ¹flipper_length_mm, ²body_mass_gHere we are filtering the penguins data set keeping only the species “Chinstrap” and those below 3.5 kilos. And we can keep going!
R
filter(penguins,
species == "Chinstrap",
body_mass_g < 3000,
sex == "male")
OUTPUT
# A tibble: 0 × 8
# … with 8 variables: species <fct>, island <fct>, bill_length_mm <dbl>,
# bill_depth_mm <dbl>, flipper_length_mm <int>, body_mass_g <int>, sex <fct>,
# year <int>
# ℹ Use `colnames()` to see all variable namesR
filter(penguins,
year >= 2008,
island == "Biscoe")
OUTPUT
# A tibble: 124 × 8
species island bill_length_mm bill_depth_mm flipper_len…¹ body_…² sex year
<fct> <fct> <dbl> <dbl> <int> <int> <fct> <int>
1 Adelie Biscoe 39.6 17.7 186 3500 fema… 2008
2 Adelie Biscoe 40.1 18.9 188 4300 male 2008
3 Adelie Biscoe 35 17.9 190 3450 fema… 2008
4 Adelie Biscoe 42 19.5 200 4050 male 2008
5 Adelie Biscoe 34.5 18.1 187 2900 fema… 2008
6 Adelie Biscoe 41.4 18.6 191 3700 male 2008
7 Adelie Biscoe 39 17.5 186 3550 fema… 2008
8 Adelie Biscoe 40.6 18.8 193 3800 male 2008
9 Adelie Biscoe 36.5 16.6 181 2850 fema… 2008
10 Adelie Biscoe 37.6 19.1 194 3750 male 2008
# … with 114 more rows, and abbreviated variable names ¹flipper_length_mm,
# ²body_mass_g
# ℹ Use `print(n = ...)` to see more rowsR
filter(penguins,
sex == "male",
species == "Chinstrap")
OUTPUT
# A tibble: 34 × 8
species island bill_length_mm bill_depth_mm flipper_l…¹ body_…² sex year
<fct> <fct> <dbl> <dbl> <int> <int> <fct> <int>
1 Chinstrap Dream 50 19.5 196 3900 male 2007
2 Chinstrap Dream 51.3 19.2 193 3650 male 2007
3 Chinstrap Dream 52.7 19.8 197 3725 male 2007
4 Chinstrap Dream 51.3 18.2 197 3750 male 2007
5 Chinstrap Dream 51.3 19.9 198 3700 male 2007
6 Chinstrap Dream 51.7 20.3 194 3775 male 2007
7 Chinstrap Dream 52 18.1 201 4050 male 2007
8 Chinstrap Dream 50.5 19.6 201 4050 male 2007
9 Chinstrap Dream 50.3 20 197 3300 male 2007
10 Chinstrap Dream 49.2 18.2 195 4400 male 2007
# … with 24 more rows, and abbreviated variable names ¹flipper_length_mm,
# ²body_mass_g
# ℹ Use `print(n = ...)` to see more rowsThe difference between & (and) and |(or)
But what if we want all the Chinstrap penguins or if body mass is below 3 kilos? When we use the comma (or the &), we make sure that all statements are TRUE. But what if we want it so that either statement is true? Then we can use the or character | .
R
filter(penguins,
species == "Chinstrap" |
body_mass_g < 3000)
OUTPUT
# A tibble: 75 × 8
species island bill_length_mm bill_depth_mm flippe…¹ body_…² sex year
<fct> <fct> <dbl> <dbl> <int> <int> <fct> <int>
1 Adelie Dream 37.5 18.9 179 2975 <NA> 2007
2 Adelie Biscoe 34.5 18.1 187 2900 fema… 2008
3 Adelie Biscoe 36.5 16.6 181 2850 fema… 2008
4 Adelie Biscoe 36.4 17.1 184 2850 fema… 2008
5 Adelie Dream 33.1 16.1 178 2900 fema… 2008
6 Adelie Biscoe 37.9 18.6 193 2925 fema… 2009
7 Adelie Torgersen 38.6 17 188 2900 fema… 2009
8 Chinstrap Dream 46.5 17.9 192 3500 fema… 2007
9 Chinstrap Dream 50 19.5 196 3900 male 2007
10 Chinstrap Dream 51.3 19.2 193 3650 male 2007
# … with 65 more rows, and abbreviated variable names ¹flipper_length_mm,
# ²body_mass_g
# ℹ Use `print(n = ...)` to see more rowsThis now gives us both all chinstrap penguins, and the smallest Adelie penguins! By combining AND and OR statements this way, we can slowly create the filtering we are after.
R
filter(penguins,
sex == "male" |
species == "Chinstrap")
OUTPUT
# A tibble: 202 × 8
species island bill_length_mm bill_depth_mm flipper_…¹ body_…² sex year
<fct> <fct> <dbl> <dbl> <int> <int> <fct> <int>
1 Adelie Torgersen 39.1 18.7 181 3750 male 2007
2 Adelie Torgersen 39.3 20.6 190 3650 male 2007
3 Adelie Torgersen 39.2 19.6 195 4675 male 2007
4 Adelie Torgersen 38.6 21.2 191 3800 male 2007
5 Adelie Torgersen 34.6 21.1 198 4400 male 2007
6 Adelie Torgersen 42.5 20.7 197 4500 male 2007
7 Adelie Torgersen 46 21.5 194 4200 male 2007
8 Adelie Biscoe 37.7 18.7 180 3600 male 2007
9 Adelie Biscoe 38.2 18.1 185 3950 male 2007
10 Adelie Biscoe 38.8 17.2 180 3800 male 2007
# … with 192 more rows, and abbreviated variable names ¹flipper_length_mm,
# ²body_mass_g
# ℹ Use `print(n = ...)` to see more rows